cg_plot_meta
cg_plot_meta.RdCreate a ggplot2 object, plotting a
user-defined metadata over time in a swimmer's plot.
ggplot2 objects retain the entirety of the provided dataset.
This allows later adjustments, such as adding extra geom_layers
with new information, or applying facets. To find this data
examine obj$data. If you save ggplot2 objects, all source
data is ALSO saved. cg_plot_meta() removes any un-used data by
default (drop_vars=TRUE). In writing a study specific
ggplot2, it is best practice to select minimal columns before
calling ggplot().
Usage
cg_plot_meta(
cg,
x = NULL,
date_dose_1 = date_dose_1,
dose_1 = dose_1,
date_dose_2 = date_dose_2,
dose_2 = dose_2,
visit = visit,
drop_vars = TRUE,
fill = NULL
)Arguments
- cg
chronogram
- x
a column of time to use as x axis. If NULL, will default to the chronogram's calendar date attribute. A user may want to derive and use alternatives eg 'daysSinceDose2'.
- date_dose_1
column containing the date of dose.
- dose_1
column containing the vaccine formulation.
- date_dose_2
column containing the date of dose.
- dose_2
column containing the vaccine formulation.
- visit
column within chronogram to indicate a study visit (i.e. samples available; NA when samples not taken, !=NA when samples available). In our small study example, serum_Ab_S fills this brief.
- drop_vars
Default TRUE. See description.
- fill
column used to determine fill
Examples
library(ggplot2)
library(patchwork)
data("built_smallstudy")
cg <- built_smallstudy$chronogram
p1 <- cg_plot_meta(cg,
visit = serum_Ab_S
)
#> Function provided to illustrate chronogram ->
#> ggplot2 interface.
#> Function assumes the
#> presence of {dose_1, date_dose_1, dose_2, date_dose_2}
#> columns.
#> Users are likely to want to write their own,
#> study-specific applications
p2 <- cg_plot(cg,
y_values = serum_Ab_S
)
#> Function provided to illustrate chronogram ->
#> ggplot2 interface.
#> Users are likely to want to write their own,
#> study-specific applications
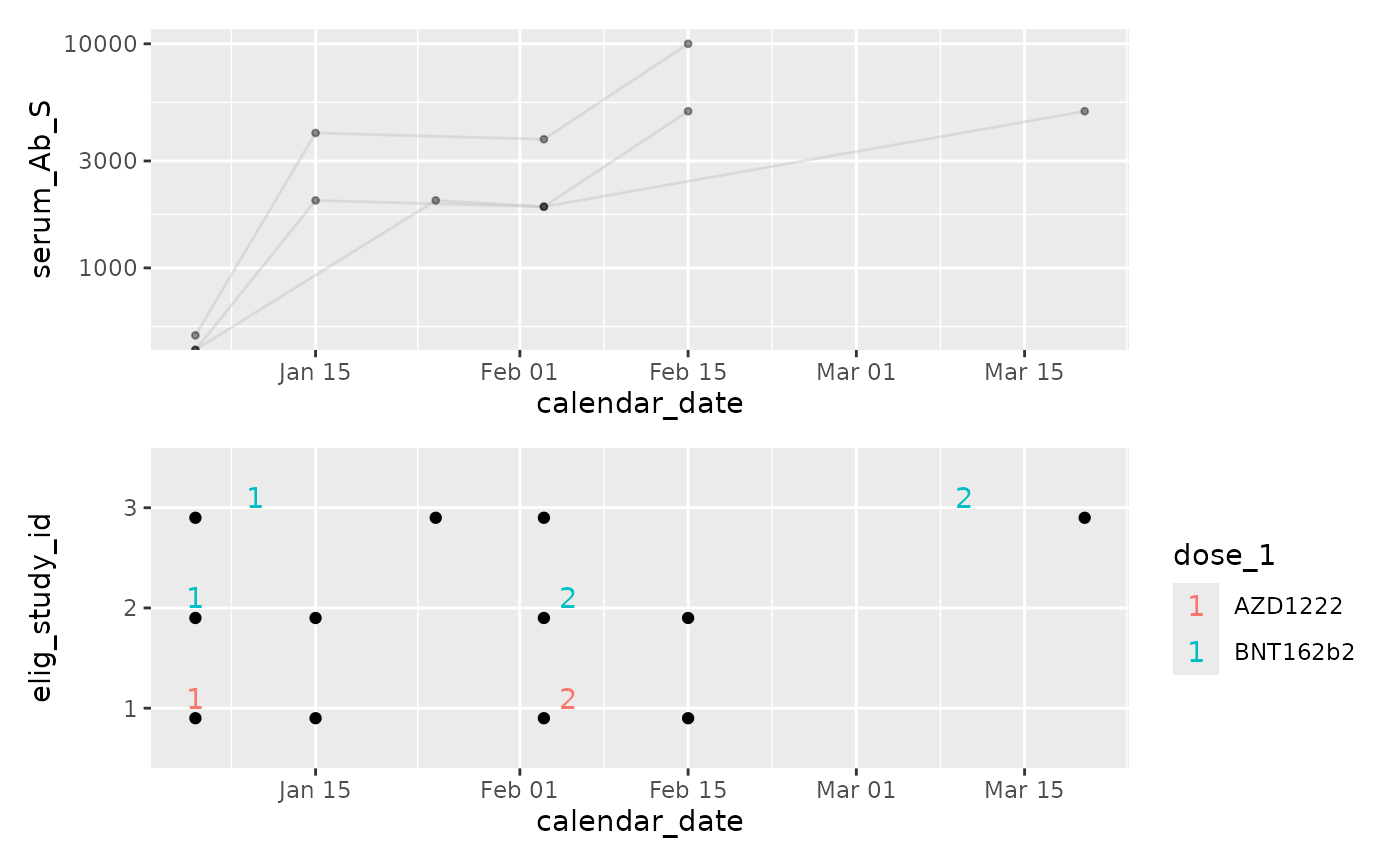
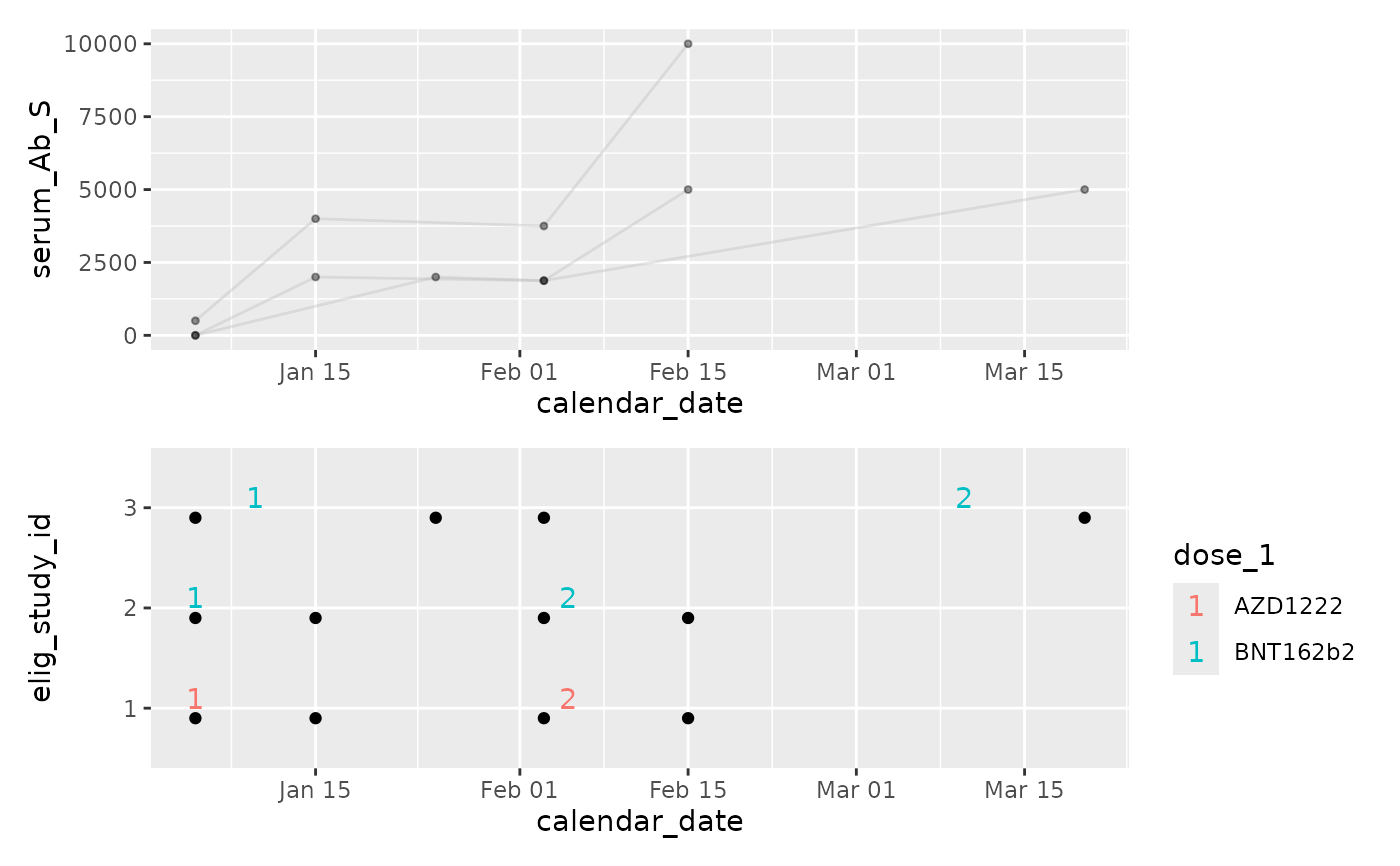
p2 / p1
 (p2 + scale_y_log10()) / p1
#> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
(p2 + scale_y_log10()) / p1
#> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.