Quickstart
chronogram.RmdThis vignette is for the impatient and showcases the features of the
chronogram package.
Construct a chronogram
Use cg_assemble() to create a chronogram from cleaned
input data.
## Fictional example data ##
data(smallstudy)
cg <- cg_assemble(
start_date = "01012020",
end_date = "10102021",
## the provided metadata ##
metadata = smallstudy$small_study_metadata,
## the column name in the metadata that contains participant IDs ##
metadata_ids_col = elig_study_id,
## column name for dates ##
calendar_date_col = calendar_date,
## the provided experiment data (we have 1 assay, so a list of 1) ##
experiment_data_list = list(smallstudy$small_study_Ab)
)
#> Checking input parameters...
#> -- checking start date 01012020
#> -- checking end date 10102021
#> -- checking end date later than start date
#> -- checking metadata
#> -- checking experiment data list
#> --- checking experiment data list slot 1
#> Input checks completed
#> Chronogram assembling...
#> -- chrongram_skeleton built
#> -- chrongram built with metadata
#> -- adding experiment data
#> --- adding experiment data slot 1 cols... elig_study_id calendar_date serum_Ab_S ...print(), glimpse() and
summary() help explore the resulting chronogram.
(View() works too, but not shown in vignette).
print(cg)
#> # A tibble: 1,947 × 10
#> # A chronogram: try summary()
#> calendar_date elig_study_id age sex dose_1 date_dose_1 dose_2 date_dose_2
#> * <date> <fct> <dbl> <fct> <fct> <date> <fct> <date>
#> 1 2020-01-01 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 2 2020-01-02 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 3 2020-01-03 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 4 2020-01-04 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 5 2020-01-05 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 6 2020-01-06 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 7 2020-01-07 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 8 2020-01-08 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 9 2020-01-09 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> 10 2020-01-10 1 40 F AZD12… 2021-01-05 AZD12… 2021-02-05
#> # ℹ 1,937 more rows
#> # ℹ 2 more variables: serum_Ab_S <dbl>, serum_Ab_N <dbl>
#> # ★ Dates: calendar_date ★ IDs: elig_study_id
#> # ★ metadata: age, sex, dose_1, date_dose_1, dose_2, date_dose_2
glimpse(cg)
#> Glimpse: chronogram
#> Dates column: calendar_date
#> IDs column: elig_study_id
#>
#> Metadata
#> Rows: 1,947
#> Columns: 6
#> $ age <dbl> 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40…
#> $ sex <fct> F, F, F, F, F, F, F, F, F, F, F, F, F, F, F, F, F, F, F, F…
#> $ dose_1 <fct> AZD1222, AZD1222, AZD1222, AZD1222, AZD1222, AZD1222, AZD1…
#> $ date_dose_1 <date> 2021-01-05, 2021-01-05, 2021-01-05, 2021-01-05, 2021-01-0…
#> $ dose_2 <fct> AZD1222, AZD1222, AZD1222, AZD1222, AZD1222, AZD1222, AZD1…
#> $ date_dose_2 <date> 2021-02-05, 2021-02-05, 2021-02-05, 2021-02-05, 2021-02-0…
#>
#> Experiment data & annotations
#> Rows: 1,947
#> Columns: 2
#> $ serum_Ab_S <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ serum_Ab_N <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
summary(cg)
#> A chronogram:
#> Dates column: calendar_date
#> IDs column: elig_study_id
#> Starts on: 2020-01-01
#> Ends on: 2021-10-10
#> Contains: 3 unique participant IDs
#> Windowed: FALSE
#> Spanning: 649 - 649 days [min-max per participant]
#> Metadata: age, sex, dose_1, date_dose_1, dose_2, date_dose_2
#> Size: 129.55 kB
#> Pkg_version: 1.0.0 [used to build this cg]Plot trajectories
Plot antibody responses over time for each individual using
cg_plot().
traj <- cg_plot(cg,
y_values = serum_Ab_S
)
#> Function provided to illustrate chronogram ->
#> ggplot2 interface.
#> Users are likely to want to write their own,
#> study-specific applications
traj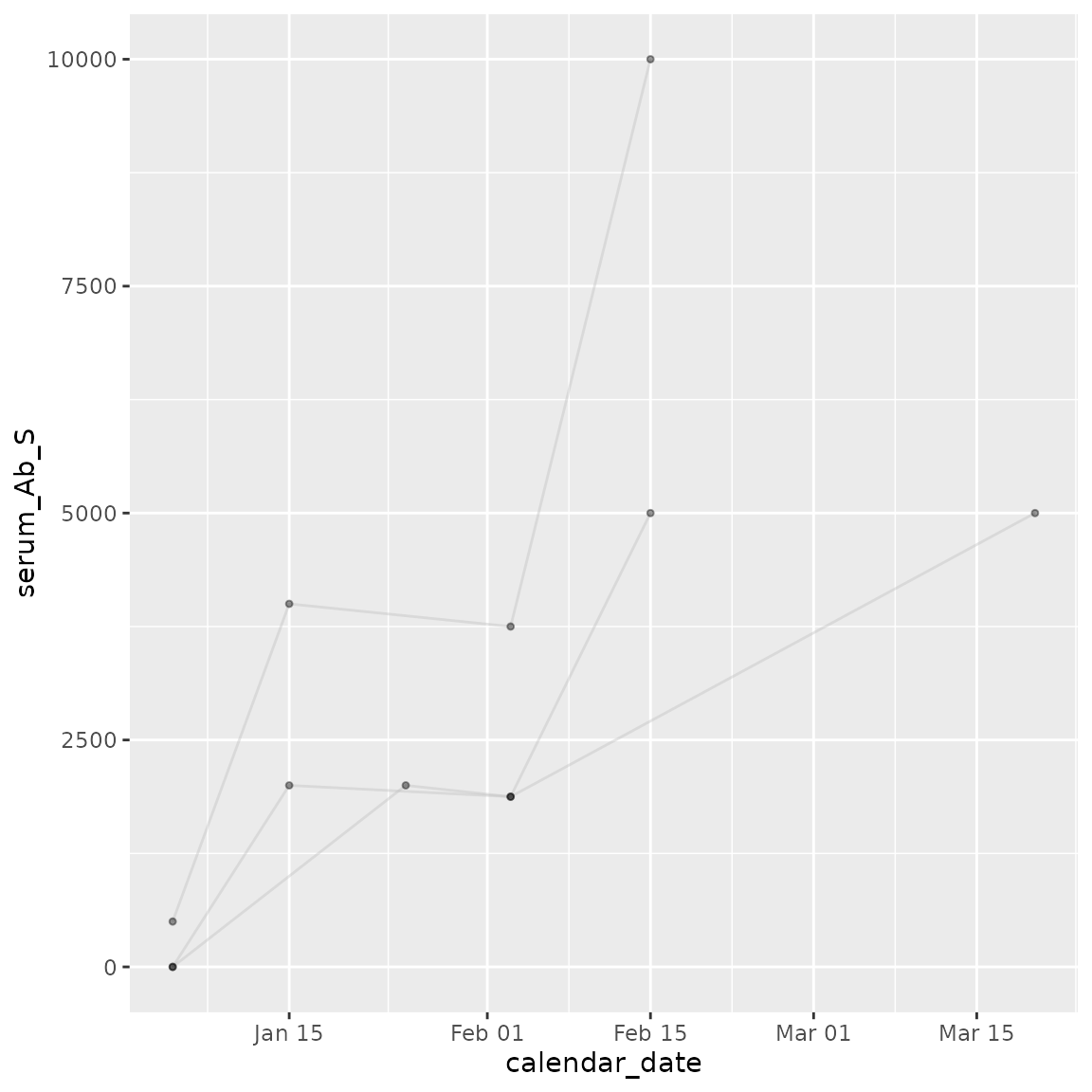
Plot a swimmers plot to visualise an individual’s sequence of events
within the study, using cg_plot_meta().
swimmer <- cg_plot_meta(cg,
visit = serum_Ab_S
)
#> Function provided to illustrate chronogram ->
#> ggplot2 interface.
#> Function assumes the
#> presence of {dose_1, date_dose_1, dose_2, date_dose_2}
#> columns.
#> Users are likely to want to write their own,
#> study-specific applications
swimmer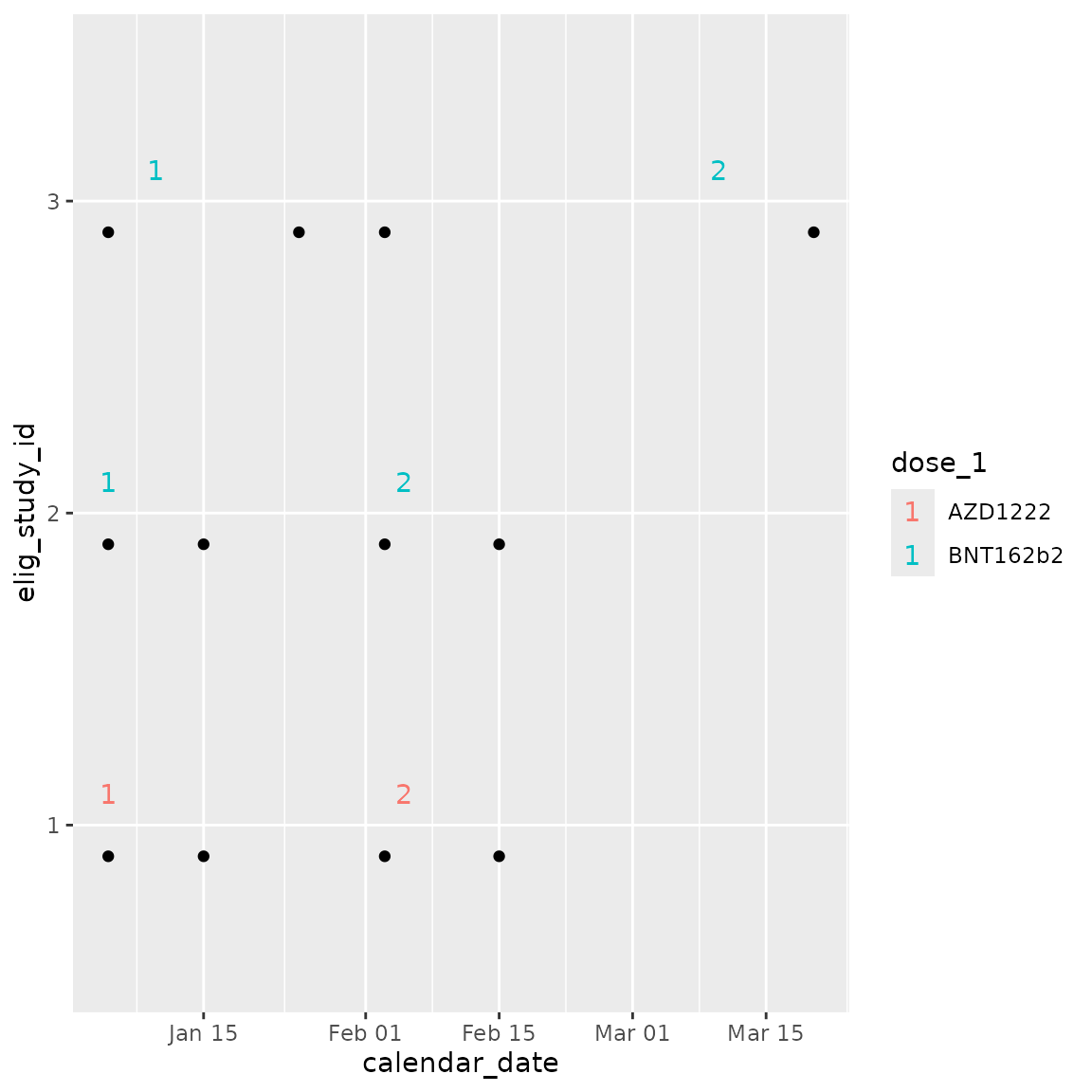
Customise plots
Using ggplot2 or patchwork
customisations.
lay <- "
a
a
a
b
"
(
(traj + labs(y = "anti-S IgG") + scale_y_log10()) /
(swimmer + theme(axis.text.y = element_blank()))
) +
plot_layout(design = lay) &
plot_annotation(tag_levels = "A") &
theme_bw(base_size = 8) &
theme(legend.position = "none") &
theme(plot.tag = element_text(size = 16, face = "bold")) &
labs(x = "date")
#> Warning in scale_y_log10(): log-10 transformation introduced infinite values.
#> log-10 transformation introduced infinite values.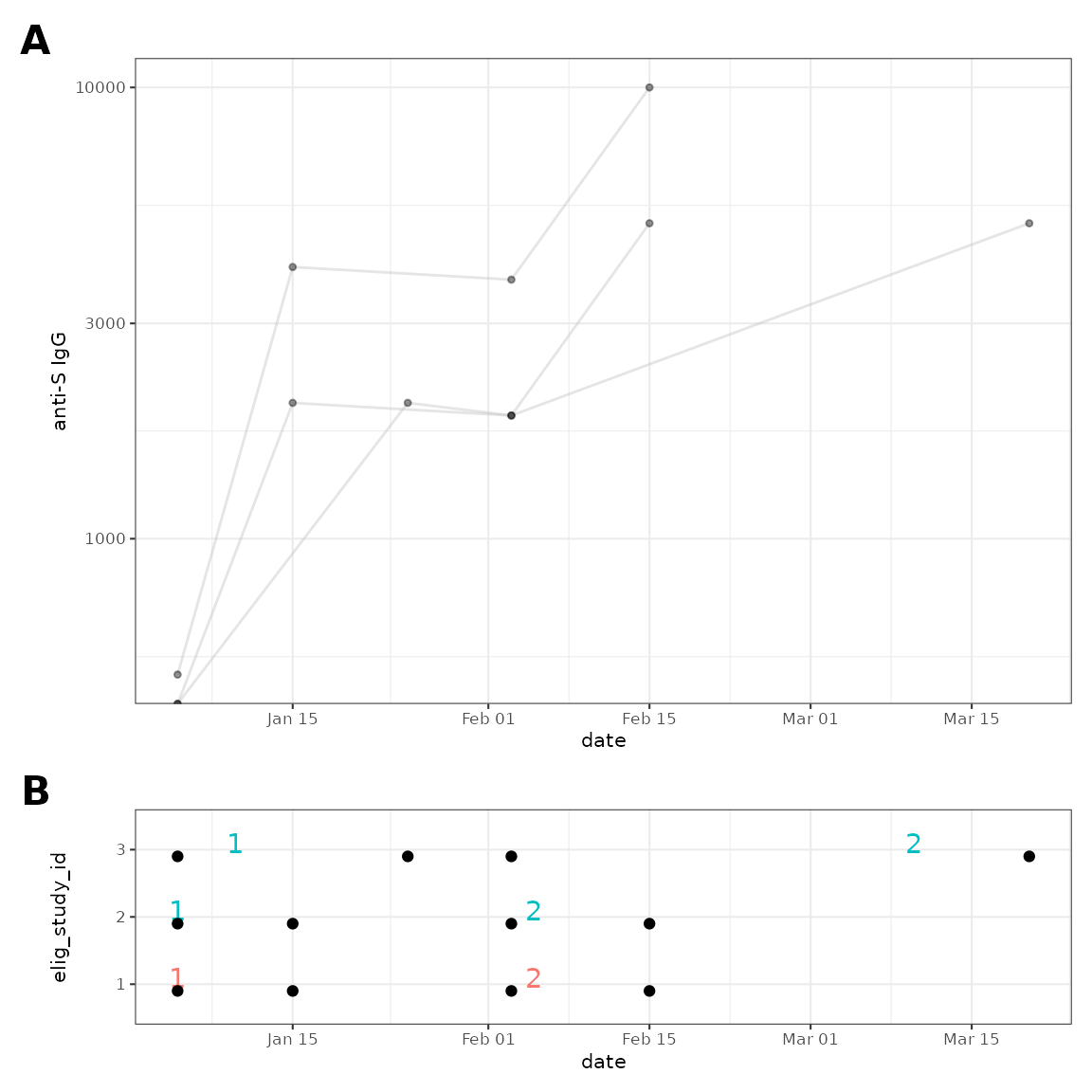
Annotate a chronogram
There are a suite of functions to find, label and count infection episodes. These aggregate across different experiment data columns, such as symptoms, PCRs, viral sequencing. See the annotation vignette.
Window a chronogram
Here, we pick dates in relation to dose 2, using
cg_window_by_metadata().
## upto 50d after dose 2 ##
cg_after_dose_2 <- cg_window_by_metadata(cg,
windowing_date_col = date_dose_2,
preceding_days = 0,
following_days = 50
)
## upto 21d before dose 2 ##
cg_before_dose_2 <- cg_window_by_metadata(cg,
windowing_date_col = date_dose_2,
preceding_days = 21,
following_days = 0
)
cg_after_dose_2 <- cg_after_dose_2 %>% mutate(cohort = "post D2")
cg_before_dose_2 <- cg_before_dose_2 %>% mutate(cohort = "pre D2")
cg_dose_2 <- bind_rows(cg_after_dose_2, cg_before_dose_2)
cg_dose_2 <- cg_dose_2 %>% mutate(
cohort =
factor(cohort,
levels = c(
"pre D2",
"post D2"
)
)
)Re-plot the windowed data
traj_2 <- cg_dose_2 %>%
cg_plot(.,
y_values = serum_Ab_S
)
#> Function provided to illustrate chronogram ->
#> ggplot2 interface.
#> Users are likely to want to write their own,
#> study-specific applications
pd <- position_dodge(0.4)
violin_2 <- cg_dose_2 %>%
## geom_line fails if lots of NA rows provided ##
filter(!is.na(serum_Ab_S)) %>%
ggplot(aes(
x = cohort, y = serum_Ab_S,
group = elig_study_id
)) +
geom_point(position = pd) +
geom_line(position = pd)
swimmer_2 <- cg_plot_meta(cg_dose_2,
visit = serum_Ab_S
)
#> Function provided to illustrate chronogram ->
#> ggplot2 interface.
#> Function assumes the
#> presence of {dose_1, date_dose_1, dose_2, date_dose_2}
#> columns.
#> Users are likely to want to write their own,
#> study-specific applications
lay2 <- "
aac
aac
aa#
bb#
"
(
(traj_2 +
labs(y = "anti-S IgG", x = "date") +
scale_y_log10()) +
(swimmer_2 +
labs(x = "date") +
theme(axis.text.y = element_blank())) +
(violin_2 +
labs(y = "anti-S IgG") +
scale_y_continuous(trans = "log10") +
annotation_logticks(sides = "l"))
) +
plot_layout(design = lay2) &
plot_annotation(tag_levels = "A") &
theme_bw(base_size = 8) &
theme(legend.position = "none") &
theme(plot.tag = element_text(size = 16, face = "bold"))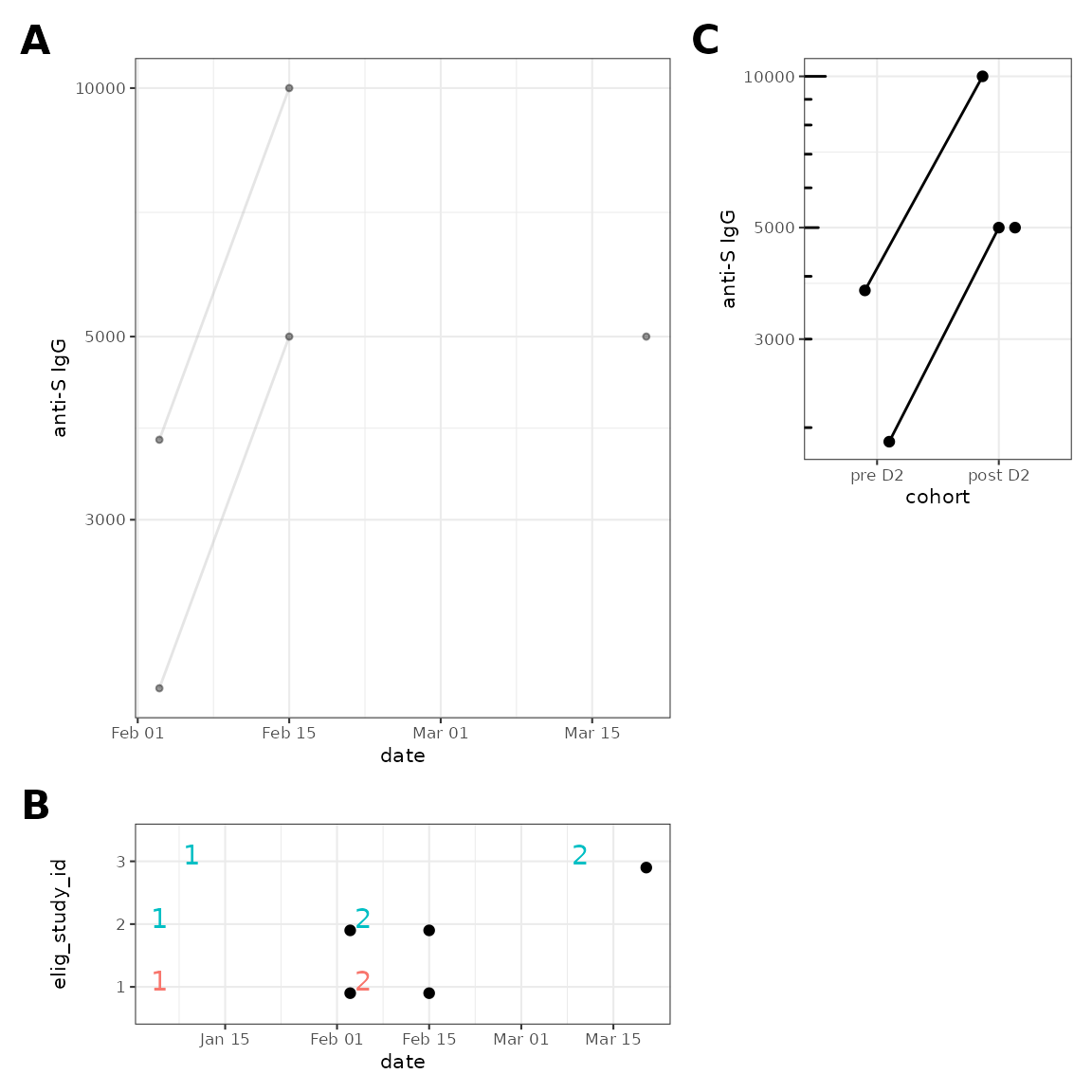
Note that for participant 3, the sample ~50d before dose 2 is now excluded.
Summary
Chronogram is now assembled and the relevant data to answer a (simple) biological question (does dose 2 boost anti-S IgG?) retrieved, plotted and readied for a statistical test.
Please see the other vignettes for a deeper demonstration of chronogram functions.
SessionInfo
sessionInfo()
#> R version 4.4.2 (2024-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.5 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] chronogram_1.0.0 patchwork_1.3.0 ggplot2_3.5.1 dplyr_1.1.4
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.6 jsonlite_1.8.9 highr_0.11 compiler_4.4.2
#> [5] tidyselect_1.2.1 tidyr_1.3.1 jquerylib_0.1.4 systemfonts_1.1.0
#> [9] scales_1.3.0 textshaping_0.4.0 yaml_2.3.10 fastmap_1.2.0
#> [13] lobstr_1.1.2 R6_2.5.1 labeling_0.4.3 generics_0.1.3
#> [17] knitr_1.48 tibble_3.2.1 desc_1.4.3 munsell_0.5.1
#> [21] lubridate_1.9.3 bslib_0.8.0 pillar_1.9.0 rlang_1.1.4
#> [25] utf8_1.2.4 cachem_1.1.0 xfun_0.49 fs_1.6.5
#> [29] sass_0.4.9 timechange_0.3.0 cli_3.6.3 pkgdown_2.1.1
#> [33] withr_3.0.2 magrittr_2.0.3 digest_0.6.37 grid_4.4.2
#> [37] lifecycle_1.0.4 vctrs_0.6.5 evaluate_1.0.1 glue_1.8.0
#> [41] farver_2.1.2 ragg_1.3.3 fansi_1.0.6 colorspace_2.1-1
#> [45] purrr_1.0.2 rmarkdown_2.29 tools_4.4.2 pkgconfig_2.0.3
#> [49] htmltools_0.5.8.1